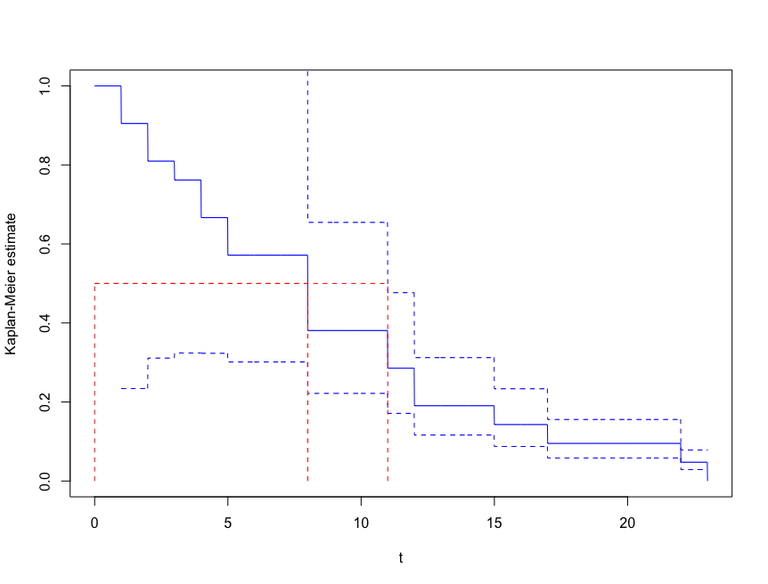
Solution: Kaplan-Meier estimator for Leukemia data
Eksempel
Here we solve problem 3.7 in ABG, using the definition in section 3.2.1 and the modifications needed to handle ties from section 3.2.2.
Solution to (a)
The Kaplan-Meier estimator for the survival function \(S(t)\) (\((3.26)\) in ABG) is given by \[\hat S(t) = \prod_{T_j \leq t} \left\{1-\frac{1}{Y(T_j)}\right\},\] where \(Y(t)\) is the number of individuals at risk just before time \(t\) and \(0 < T_1 < \ldots\) are the event times. Here we have ties, since the remission time is rounded to whole weeks. We therefore adjust the estimates using the correction \((3.32)\) in ABG, given by \[\hat S(t) = \prod_{T_j \leq t} \left\{1-\frac{d_j}{Y(T_j)}\right\},\] where \(d_j\) is the number of events happening at time \(T_j\).
For the placebo group we have

Code 1 contains some R code that calculates and plots the Kaplan-Meier estimate for the placebo group according to the formula stated above, with confidence limits from \((3.30)\).
Times = c(0,1,2,3,4,5,8,11,12,15,17,22,23) #Event times
d = c(0,2,2,1,2,2,4,2,2,1,1,1,1) #Number of events
c = rep(0,length(Times)) #Censored observations
Kaplan_Meier = function(Times,d,c){
# Calculates the Kaplan-Meier estimate according to 3.32 in ABG
m = length(Times)
Y = rep(21,m)
Y[2:m] = Y[2:m] - cumsum(d+c)[1:(m-1)]
S = cumprod(1 - d/Y)
Shat = stepfun(Times[2:m],S)
return(Shat)
}
SigmaHat2 = function(Times,d,c){
# Calculates the variance estimate in 3.14/3.15,
# which will then be used to calculate the estimated variance
# of the Kaplan-Meier estimates
m = length(Times)
Y = rep(21,m)
Y[2:m] = Y[2:m] - cumsum(d+c)[1:(m-1)]
Deltahatsigma2 = rep(0,m)
for (j in 1:m){
# This is the adjusted inner sum
if (d[j] > 0){
for (l in 0:(d[j]-1)){
Deltahatsigma2[j] = Deltahatsigma2[j] + 1/(Y[j]-l)^2
}
}
}
hatsigma2 = stepfun(Times[2:m],cumsum(Deltahatsigma2))
return(hatsigma2)
}
hatS = Kaplan_Meier(Times,d,c)
t = seq(0,max(Times),by=0.01)
hatTau2 = hatS(t)^2*SigmaHat2(Times,d,c)(t)
exponent = qnorm(0.975)*sqrt(hatTau2)/(hatS(t)*log(hatS(t)))
conf_upper = hatS(t)*exp(-exponent)
conf_lower = hatS(t)*exp(exponent)
par(mfrow = c(1,1))
plot(t,hatS(t),type="l",col="blue",ylab="Kaplan-Meier estimate")
lines(t,conf_upper,lty=2,col="blue")
lines(t,conf_lower,lty=2,col="blue")
Figure 1 shows the resulting plot:
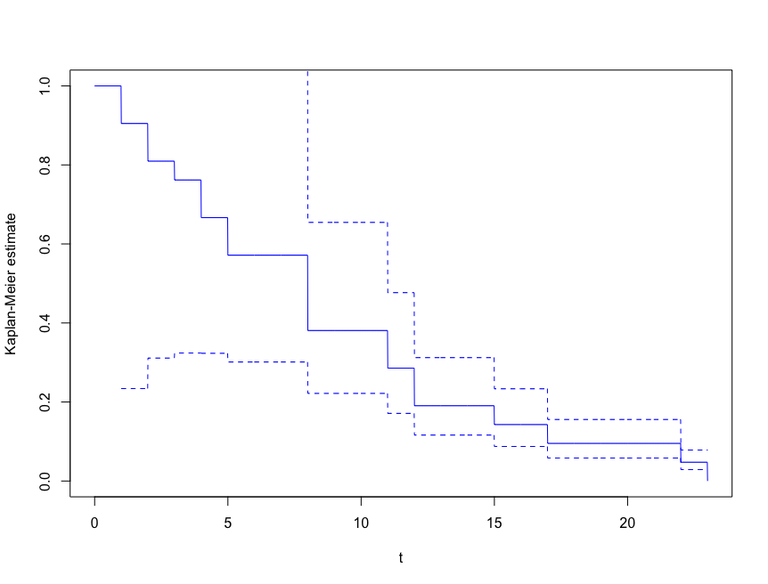
Solution to (b)
We use the method described in section 3.2.3 to find an estimate and a \(95\%\) confidence interval for the median relapse time for the placebo group. From the plot in figure 1 we see that the estimate jumps below 0.5 at \(t=8\), and hence our estimate is \(\hat \xi_{0.5} = 8\). For the confidence interval, first observe that the lower confidence bound for the Kaplan-Meier estimate is always smaller than 0.5, and so the lower bound for our confidence interval for the median relapse time is 0. The upper confidence bound jumps below 0.5 at \(t=11\), and hence our confidence interval is \([0,11]\). Figure 2 illustrates this: